Predicting Additive Release from Plastics in the End-of-Life Phase
Background: Plastic production, usage, and waste have increased and continue to increase significantly every year. What many people do not realize, however, is that there is more to plastic than meets the eye: plastic used for everything from medical syringes and bottles to bags and wrappers contains more than just its polymer (PET, HDPE, PVC, etc.). Plastic often contains chemical additives that give that plastic its unique properties. Plasticizers, for instance, make the plastic more flexible, flame retardants make it fire-resistant, and blowing agents make it foamy. These additives can cause a variety of problems. Because each formulation is different (even the plastic used for bottles may differ between water and soda brands), the mixture of additives can make recycling, especially the conventional method of mechanical recycling, difficult. Besides that, the additives in plastic can migrate out of the plastic matrix because they are not chemically bound to the polymers, especially when plastic products are left for long periods of time, like they are in landfills or when dropped as litter.
This project aims to estimate the additive migrations, or releases, of plastic with a focus on the end-of-life phase. Based on municipal solid waste data collected and published by the US EPA every year (including information like total amount of waste generated, sent to landfills, recycled, incinerated, etc.) and different literature sources (used to determine additive types and identities, migration, leaching, etc.), a mass balance was carried out on the life cycle of plastic to predict the additive releases in a given year.

Sankey diagram showing plastic flow through its life cycle, from manufacturing to various end-of-life options.
A software tool, known as EoLPAFT (End-of-Life Plastic Additives Flow Tracker), was developed in Python. This program allows users to generate their own mass balance using their own municipal solid waste (MSW) data. EoLPAFT was designed with a non-technical policymaker-user in mind and the goal is that any individual or decision-making entity (from small-town waste managers to the EPA) can use this tool to determine the effects of different policies on their waste stream and the resulting migration of additives into the environment.

A visual representation of EoLPAFT and the capabilities it provides.
The tool can be accessed and downloaded at this link. A paper with more information on the tool’s development is currently under review.
Solvent Recovery for Reuse and Recycling
Background: Solvents are mainly used in the pharmaceutical, fine, and specialty chemicals industries for cleaning, extraction, dissolution, and as reaction media. Thus, they are also a major constituent in the waste streams of these industries. Incineration, on-, and off-site disposal are the traditional solvent waste handling methods. These conventional disposal techniques increase the carbon and environmental footprint of industrial processes. Instead of disposal, there can be a more preferable way to recover these solvents for reuse. Therefore, having a framework to help recover solvents for reuse will help reduce the cost and environmental footprint of these production processes.
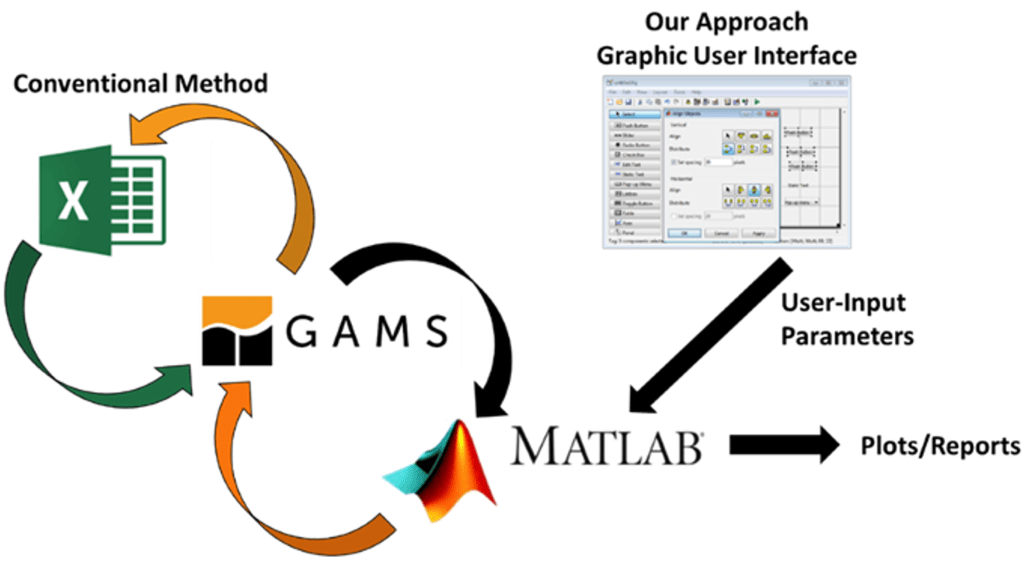
Proposed Solution and Projections: To help recover solvents from industrial waste streams, a proposed framework leading to a software application was developed that has the potential to help industries save capital and reduce their environmental footprint.

Computational Tool: Through support from the U.S. Environmental Protection Agency Pollution Prevention program, we have developed a computational tool to help industries find potential optimal pathways to recover solvents from waste streams for reuse. Through this tool, industries can estimate the annual capital and operating costs and reduce life cycle emissions when solvent recovery is implemented. Comparative assessment can be made with conventional disposal technique such as incineration. The standalone tool can be accessed on GitHub.
More information about the development can be found in the publication:
Management of IBS (Irritable Bowel Syndrome)
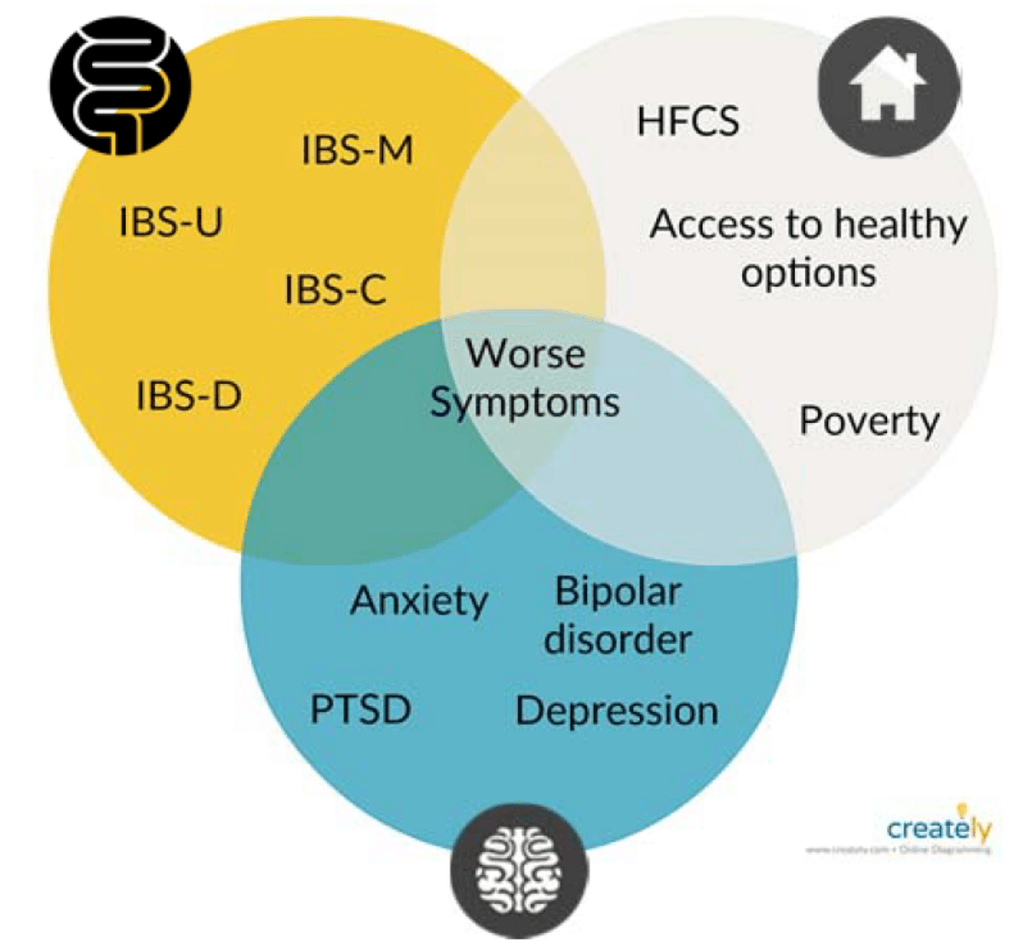
IBS is a common gastrointestinal disorder affecting the large intestine. It is estimated to affect 10 to 15 percent of the world’s population and 25 to 45 million people in the US alone. Symptoms of IBS include cramping, abdominal pain, bloating, constipation, diarrhea, or both. There are no definitive causes for IBS nor is there a definitive way to diagnose patients with IBS. Doctors base their diagnosis on patient-doctor discussions and the elimination of other possible conditions. This long and often tedious process is a reason why this research group sought out to see if any specific conditions within the body were consistent for certain subtypes of IBS, specifically the microbiome of the gut.

This research utilized random forest modeling to find correlations within data provided by Cooper Medical School to accurately diagnose patients with IBS based on their gut microbiome. The random forest model, made up of multiple decision trees, determined the top six gut bacteria found to be most important in determining a patient’s subtype of IBS. This will allow for healthcare clinicians to provide the best diets and supplements as studies have proven that the diet of a patient has a large influence on the symptoms a patient experiences as well as their severity.

With this research, an online application was developed where the user can input their own values for these top 6 gut bacteria and get a prediction for the subtype of IBS the patient could have. The figure below depicts what the user will see when values are inputted and a prediction is made.
The IBS Diagnosis Application can be accessed publicly at https://ibsdiagnosis.shinyapps.io/IBSDiagnosis/
A tutorial on how to use the IBS Diagnosis Application is provided below.
